The Table Access Protocol (TAP) is a way to access astronomical databases using the Astronomical Data Query Language (ADQL). The ADQL is a variant of SQL that supports querying grammar for astronomical operations.
Simbad is a database for astronomical objects that is updated constantly from the literature. It can be accessed by various methods including TAP.
I recently wanted to
- retrieve all related bibliography for a set of objects
- get all radial velocity measurements for a set of objects
It turns out this is fairly simple with pyvo and ADQL.
Pyvo provides a collection of python modules related to Virtual observatory. For Simbad, we can simply create an instance of its vo.dal.TAPService class with the baseurl set to TAP endpoint.# import libraries
%
pyvo.__version__='1.3'
=
Let's see all the available tables.
# get all tables
# tables method of TAPService parses the response of /tables,
# but it will result in exception due to a possible bug in astropy vo
# but you can also just query the database for this info
=
number of tables available = 29
| table_name | description |
|---|---|
| object | object |
| basic | General data about an astronomical object |
| ids | all names concatenated with pipe |
| alltypes | all object types concatenated with pipe |
| otypedef | all names and definitions for the object types |
| ident | Identifiers of an astronomical object |
Get all velocity measurements
The relevant table for RV measurements is mesVelocities. We also need ident table which has two columns, oidref and id where oidref is unique internally, and id is all possible known names of sources. we use oidref to crossmatch tables. We can first check what columns are in mesVelocities and ident tables.
"""Pretty print column description table"""
=
=
return
| colname | dtype | description |
|---|---|---|
| str6 | str3 | str26 |
| id | |O | Identifier |
| oidref | <i8 | Object internal identifier |
| colname | dtype | description |
|---|---|---|
| str14 | str3 | str71 |
| bibcode | |O | measurement bibcode |
| d | <U1 | 'D' if the resolution is a conversion from the dispersion |
| meanError | <f4 | sigma(Value) |
| meanError_prec | <i2 | Precision (# of decimal positions) associated with the column meanError |
| mespos | <i2 | Position of a measurement in a list of measurements |
| nature | |O | nature of the measurement |
| nbmes | <i2 | Number of measurements |
| ... | ... | ... |
| remark | <U1 | colon is uncertain question mark is questionable |
| remarks | |O | Remarks |
| resolution | <i4 | Resolution |
| velType | |O | velocity type (v, z or cz) |
| velValue | <f8 | Velocity |
| velValue_prec | <i2 | Precision (# of decimal positions) associated with the column velValue |
| wdomain | |O | Wavelength domain (Rad,mm,IR,Opt,UV,XRay,Gam) |
We will query stars by their HIP or TYC identifiers. For this, we make a table with one column (name), and upload this in order to crossmatch in VOTable format.
=
Grab oidref from ident table, and query mesVelocities.
=
%%
=
CPU times: user 178 ms, sys: 11 ms, total: 189 ms
Wall time: 3.56 s
Even for about 10k stars, this is very fast. We can access astropy.table.Table instance of the result as result.table. Better yet, because I know there are multiple measurements for some stars, I can convert it to pandas.DataFrame for further inspection.
=
# decode all bytestrings
=
quality
count 744
unique 438
top TYC 2342-544-1
freq 14
A count 508
unique 360
top HIP 107362
freq 6
B count 864
unique 709
top TYC 6166-1405-1
freq 4
C count 382
unique 313
top TYC 9344-293-1
freq 3
D count 89
unique 84
top TYC 8966-1241-1
freq 3
E count 31
unique 30
top HIP 109388
freq 2
Name: name, dtype: object
I see that there are quite a few stars with multiple quality A measurements. Let's see if they agree.
=
# see if multiple quality A measurements agree with each other
# make sure bibcode for the measurements are different
=
96
# undersample for presentation
;
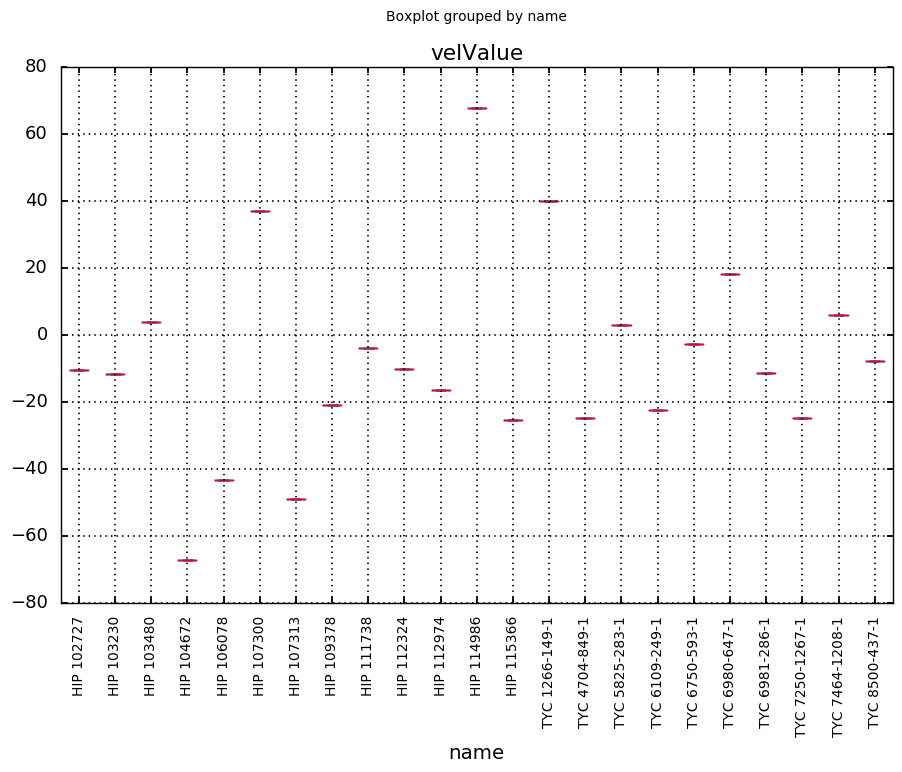
..and they do for the most part!
Get all bibliographies
Same goes for querying all bibliographies related to a set of objects. This time, we need to query ref and also need has_ref in order to crossmatch oidref with oidbibref, which is cross-identified with ref.oidbib.
<Table length=10>
| colname | dtype | description |
|---|---|---|
| str9 | str3 | str28 |
| bibcode | |O | Bibcode |
| doi | |O | DOI designation |
| journal | |O | Abbreviation for the journal |
| last_page | <i4 | Last page number |
| nbobject | <i4 | Number of objects studied in |
| oidbib | <i8 | Bibcode internal identifier |
| page | <i4 | page number |
| title | |O | Title |
| volume | <i4 | volume number |
| year | <i2 | Publication year |
<Table length=5>
| colname | dtype | description |
|---|---|---|
| str10 | str3 | str33 |
| obj_freq | <i2 | flag |
| oidbibref | <i8 | Bibcode internal identifier |
| oidref | <i8 | Object internal identifier |
| ref_flag | <i2 | flag |
| ref_raw_id | |O | id like it appears in the article |
=